Edward S. Boyden, Ph.D.
AIMBE College of Fellows Class of 2018 For breakthrough contributions to neuroengineering, including the development of optogenetics and expansion microscopy for mapping and controlling the brain.
A new way to see the activity inside a living cell
Via MIT News | November 28, 2023Using fluorescent labels that switch on and off, MIT engineers can study how molecules in a cell interact to control the cell’s behavior.
Living cells are bombarded with many kinds of incoming molecular signal that influence their behavior. Being able to measure those signals and how cells respond to them through downstream molecular signaling networks could help scientists learn much more about how cells work, including what happens as they age or become diseased.
Right now, this kind of comprehensive study is not possible because current techniques for imaging cells are limited to just a handful of different molecule types within a cell at one time. However, MIT researchers have developed an alternative method that allows them to observe up to seven different molecules at a time, and potentially even more than that… Continue reading.
40 Hz vibrations reduce Alzheimer’s pathology, symptoms in mouse models
Via MIT | June 5, 2023Evidence that noninvasive sensory stimulation of 40 Hz gamma frequency brain rhythms can reduce Alzheimer’s disease pathology and symptoms, already shown with light and sound by multiple research groups in mice and humans, now extends to tactile stimulation. A new study by MIT scientists shows that Alzheimer’s model mice exposed to 40 Hz vibration for an hour a day for several weeks showed improved brain health and motor function compared to untreated controls.
The MIT group is not the first to show that gamma frequency tactile stimulation can affect brain activity and improve motor function, but they are the first to show that the stimulation can also reduce levels of the hallmark Alzheimer’s protein phosphorylated tau, keep neurons from dying or losing their synapse circuit connections, and reduce neural DNA damage… Continue reading.
Self-assembling proteins can store cellular “memories”
Via MIT | January 2, 2023As cells perform their everyday functions, they turn on a variety of genes and cellular pathways. MIT engineers have now coaxed cells to inscribe the history of these events in a long protein chain that can be imaged using a light microscope.
Cells programmed to produce these chains continuously add building blocks that encode particular cellular events. Later, the ordered protein chains can be labeled with fluorescent molecules and read under a microscope, allowing researchers to reconstruct the timing of the events… Continue reading.
A deep dive into cells’ RNA reality
Via EurekAlert | March 17, 2021Human cells typically transcribe half of their roughly 20,000 genes into RNA molecules at any given time. Just like with proteins, the function of those RNA species not only relies on their abundance but also their precise localization within the 3D space of each cell. Many RNA molecules convey gene information from the cell’s nucleus to the protein-synthesizing machinery distributed throughout the cytoplasm (messenger RNAs or mRNAs), others are components of that machinery itself, while still different ones regulate genes and their expression, or have functions that remain to be discovered. Importantly, many diseases including cancer and neurological diseases have signatures that appear as changes in the abundance and distribution of RNAs.
To enable the analysis of a cells’ complete collection of RNAs known as their transcriptome in their 3D space (spatial transcriptomics), Wyss Institute synthetic biologists led by Core Faculty member George Church, Ph.D. in 2014 reported FISSEQ, an impactful spatial sequencing technology that is able to simultaneously read the sequences of thousands of those RNAs and visualize their three-dimensional coordinates. However, FISSEQ’s powerful ability to sequence this large number of RNA targets on-location comes at a price: its detection efficiency and sensitivity for many of them is relatively low, especially when their expression is low to start with or dialed down in disease… Continue reading.
Imaging method reveals a “symphony of cellular activities”
Via MIT | November 23, 2020Within a single cell, thousands of molecules, such as proteins, ions, and other signaling molecules, work together to perform all kinds of functions — absorbing nutrients, storing memories, and differentiating into specific tissues, among many others.
Deciphering these molecules, and all of their interactions, is a monumental task. Over the past 20 years, scientists have developed fluorescent reporters they can use to read out the dynamics of individual molecules within cells. However, typically only one or two such signals can be observed at a time, because a microscope cannot distinguish between many fluorescent colors.
MIT researchers have now developed a way to image up to five different molecule types at a time, by measuring each signal from random, distinct locations throughout a cell. This approach could allow scientists to learn much more about the complex signaling networks that control most cell functions, says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology and a professor of biological engineering, media arts and sciences, and brain and cognitive sciences at MIT… Continue reading.
Ed Boyden elected to National Academy of Sciences
Via MIT | May 1, 2019Ed Boyden has been elected to join the National Academy of Sciences (NAS). The organization, established by an act of Congress during the height of the Civil War, was founded to provide independent and objective advice on scientific matters to the nation, and is actively engaged in furthering science in the United States. Each year NAS members recognize fellow scientists through election to the academy based on their distinguished and continuing achievements in original research.
“I’m very honored and grateful to have been elected to the NAS,” says Boyden. “This is a testament to the work of many graduate students, postdoctoral scholars, research scientists, and staff at MIT who have worked with me over the years, and many collaborators and friends at MIT and around the world who have helped our group on this mission to advance neuroscience through new tools and ways of thinking… Continue reading.
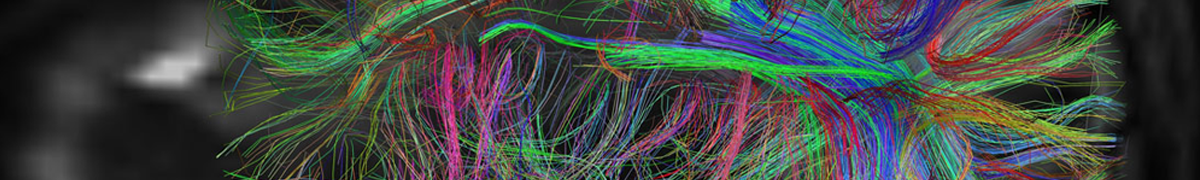
Mapping the brain at high resolution
Via MIT | January 17, 2019Researchers have developed a new way to image the brain with unprecedented resolution and speed. Using this approach, they can locate individual neurons, trace connections between them, and visualize organelles inside neurons, over large volumes of brain tissue.
The new technology combines a method for expanding brain tissue, making it possible to image at higher resolution, with a rapid 3-D microscopy technique known as lattice light-sheet microscopy. In a paper appearing in Science Jan. 17, the researchers showed that they could use these techniques to image the entire fruit fly brain, as well as large sections of the mouse brain, much faster than has previously been possible. The team includes researchers from MIT, the University of California at Berkeley, the Howard Hughes Medical Institute, and Harvard Medical School/Boston Children’s Hospital.
This technique allows researchers to map large-scale circuits within the brain while also offering unique insight into individual neurons’ functions, says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology, an associate professor of biological engineering and of brain and cognitive sciences at MIT, and a member of MIT’s McGovern Institute for Brain Research, Media Lab, and Koch Institute for Integrative Cancer Research… Continue reading.
Team invents method to shrink objects to the nanoscale
Via MIT | December 13, 2018MIT researchers have invented a way to fabricate nanoscale 3-D objects of nearly any shape. They can also pattern the objects with a variety of useful materials, including metals, quantum dots, and DNA.
“It’s a way of putting nearly any kind of material into a 3-D pattern with nanoscale precision,” says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology and an associate professor of biological engineering and of brain and cognitive sciences at MIT.
Using the new technique, the researchers can create any shape and structure they want by patterning a polymer scaffold with a laser. After attaching other useful materials to the scaffold, they shrink it, generating structures one thousandth the volume of the original… Continue reading.
Dr. Edward Boyden Inducted into Medical and Biological Engineering Elite
Via AIMBE | April 10, 2018WASHINGTON, D.C.—The American Institute for Medical and Biological Engineering (AIMBE) has announced the induction of Edward S. Boyden, Ph.D., Associate Professor, Media Lab and McGovern Institute, Departments of Biological Engineering and Brain and Cognitive Sciences, and Co-Director, Center for Neurobiological Engineering, Massachusetts Institute of Technology, to its College of Fellows. Dr. Boyden was nominated, reviewed, and elected by peers and members of the College of Fellows for breakthrough contributions to neuroengineering, including the development of optogenetics and expansion microscopy for mapping and controlling the brain.
 AIMBE
AIMBE
